6 Output File Format Specification
The DS-PAW 2023A version’s default output files in JSON format can be directly analyzed and processed using Device Studio. Additionally, the output files now support the hdf5 format. You can download vitables (run pip install vitables in a Python environment) or HDFView to view the hdf5 format files, and utilize the python scripts provided in Auxiliary Tool User Guide for result analysis.
Except for the charge density file rho.h5 and the solvent-bound charge density output file rhoBound.h5, the filenames of other output files depend on the task type. Currently, DS-PAW supports 14 task types, and the corresponding h5 filenames are: relax.h5, scf.h5, band.h5, dos.h5, potential.h5, elf.h5, pcharge.h5, frequency.h5, elastic.h5, neb.h5, phonon.h5, aimd.h5, epsilon.h5, and wannier.h5.
DS-PAW 2023A currently supports output files in .json format, but users are not advised to continue using this format for analysis. DS-PAW will completely remove the json format output during iterative versions, ceasing maintenance and updates for this format. Users can control whether to output json files via the io.outJsonFile parameter.
6.1 relax.h5
relax.h5 is the output file when task = relax; this file is not output when the task type is something else.
The relax.h5 file contains at least 9 basic structures:

AtomInfo saves the basic structural information of the system, such as cell size, atomic positions, etc.;

Eigenvalue stores the number of bands calculated, spin information, the number of k-points and their coordinates, the orbital occupation numbers and energy eigenvalues of each band at each k-point;

Electron saves the total number of valence electrons in the system;
Energy stores the total energy and Fermi energy;
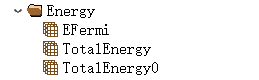
The force on each atom during the relaxation process is saved in Force;
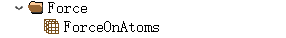
MagInfo stores the total magnetic moment information of atoms; if projections are enabled, it stores the projected magnetic moment information.
The RelaxInfo saves the stress and pressure data of the system at each step during structural relaxation;

Stress stores the stress magnitude in each direction of the unit cell, and the system pressure.

Structures stores structure and magnetic moment data during relaxation;

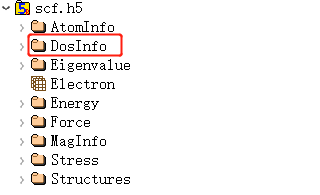
6.2 scf.h5
scf.h5 is the output file when task = scf; this file is not output for other task types.
The scf.h5 file contains at least 8 fundamental structures, with basic information consistent with relax.h5:

Under *task = scf, calculations for various functionalities can be controlled through parameters such as sys and io. The generated scf.h5 file will store data corresponding to these functionalities. Specifically, the calculations can be categorized as follows:
By setting
io.optical = true, linear optical properties are calculated based on the self-consistent field (SCF) calculation:



Calculate Bader charge based on the self-consistent calculation by setting
io.bader = true:


Perform ferroelectric calculations based on the self-consistent calculation by setting
io.polarization = true:


Perform fixed potential calculations in the self-consistent calculation by setting
sys.fixedP = true:
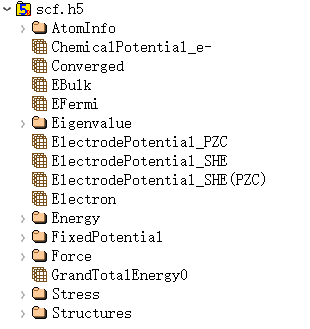
where ChemicalPotential_e is the electronic chemical potential of the system; EBulk is the negative value of the Fermi level shift under the implicit solvent model; ElectrodePotential gives the potential value under different calibration standards; and GrandTotalEnergy0 gives the total energy of the system under the grand canonical ensemble of electrons.
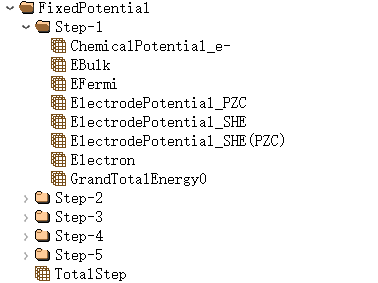
Expand under the fixedPPotential tag to summarize information on key parameters during the electronic iteration.
- By setting
io.band = true,io.dos = true,io.potential = true, andio.elf = true, Perform band structure calculation, density of states calculation, potential function calculation, and electron localization density calculation based on the self-consistent calculation:
- By setting



6.3 rho.h5
rho.h5 is the charge density output file for each task.
rho.h5 contains two structures:

Where AtomInfo is consistent with the AtomInfo structure in the relax.h5 file, and Rho stores the charge density data:

6.4 rhoBound.h5
rhoBound.h5 is the output file for the solvent-bound charge density in the solvation model.
rhoBound.h5 contains two structures:

Where AtomInfo is largely consistent with the AtomInfo structure in the relax.h5 file, and Rho stores the solvent-bound charge density data:
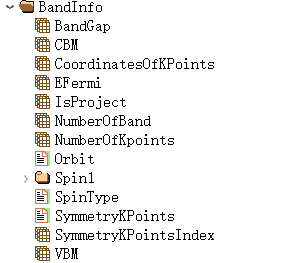
6.5 band.h5
band.h5 is the output file for each task = band. This file is not output when the task type is different.
band.h5 contains at least 3 structures:

The structures of AtomInfo, Structures, and the relax.h5 file are consistent. BandInfo stores the band structure data:
The band folding calculation corresponding to band.h5 should contain at least 4 structures:

The structure of the structs corresponding to AtomInfo, Structures, and the relax.h5 file are consistent. BandInfo stores band data, and UnfoldingBandInfo stores band unfolding data.

6.6 dos.h5
dos.h5 is the output file for task = dos; this file is not output when the task type is different.
dos.h5 contains at least 3 structures:

Among them, the structure of the structures corresponding to AtomInfo, Structures, and the file relax.h5 are consistent, and DosInfo stores the density of states data:

6.7 potential.h5
potential.h5 is an output file under task = potential. This file is not generated for other task types.
The potential.h5 file contains at least 3 structures:
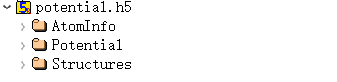
Where Potential stores the potential function data:

6.8 elf.h5
elf.h5 is the output file for task = elf. When the task type is different, this file is not output.
elf.h5 contains at least 3 structures:

Where elf stores the local density data:

6.9 pcharge.h5
pcharge.h5 is the output file under task = pcharge; this file is not output when the task type is different.
The pcharge.h5 file contains two structures:
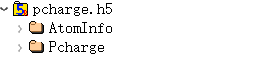
Here, Pcharge stores partial charge density data:

6.10 optical.h5
optical.h5 is the output file for task = optical; this file is not generated for other task types.
optical.h5 contains four structures:

Basic structural information of the system, such as unit cell size and atomic positions, is stored in AtomInfo:

The opticalInfo variable stores data on various properties from optical calculations:

The optical calculation structure information is saved in Structures:

WaveDerivate stores the derivative array of wave functions with respect to k-points, with a size of: (real part, imaginary part) * NumberOfBands (after selection) * NumberOfKPoints * NumberOfSpin * (x, y, z); DerivativeIndex gives the dimension of the derivative array; DerivativeValue gives the value of the derivative array.

6.11 frequency.h5
frequency.h5 is the output file for task = frequency; this file is not generated for other task types.
With spin considered, frequency.h5 contains four structures:

Where frequency data is stored in FrequencyInfo:

6.12 elastic.h5
elastic.h5 is the output file for task = elastic. This file is not output when the task type is other than elastic.
With spin considered, elastic.h5 contains four structures:
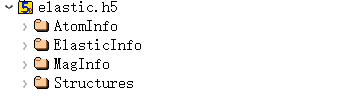
Elastic data is stored in ElasticInfo:

6.13 neb.h5
neb.h5 is the output file in the top-level directory when task = neb.
neb.h5 contains five structures:

The BarrierInfo stores the maximum force, reaction coordinate (reaction distance between each image and the initial 00 structure), maximum shear force, and total energy data:
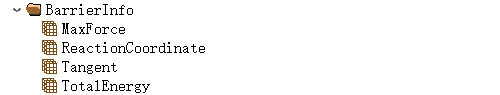
Where the switch for saving the initial and final states of the calculation is stored in IniFin:
The LoopInfo object stores the energy and force changes for each image during the neb optimization process:

In which RelaxedStructure stores the structure data after optimization of each image:

Where UnrelaxStructure stores the structural data before optimization of each image:

6.14 neb01.h5
neb01.h5 is the output file in the 01 subdirectory when task = neb. Similarly, the neb02.h5 file will be generated in the 02 subdirectory.
In the spin-unpolarized case, neb01.h5 contains 12 structures:

Among them, the structures of AtomInfo, Eigenvalue, Electron, Energy, Force, Stress, and Structures are consistent with the corresponding structure in the relax.h5 file;
In the Distance data, the distance change of the reaction atom between the initial and final images during the optimization process is stored.

where MaxForce stores the maximum force data for image 1 during optimization;
Where NebSize stores the maximum number of steps in the transition state calculation.
where Tangent stores the data of the change in the tangent force of image 1 during the optimization process;
where TotalEnergy stores the total energy change data of image 1 during the optimization process;
6.15 phonon.h5
phonon.h5 is the output file under task = phonon. This file is not output when the task type is different.
When
phonon.method = dfpt, the phonon.f5 file is as follows:
When the dfpt method is used to calculate phonon band structure and density of states, it enables the calculation of dielectric constants and phonon thermodynamics. The phonon.h5 file contains 9 structures:

Where BandInfo and DosInfo respectively store the band structure and density of states data, their structures are consistent with the corresponding structures in band.h5 and dos.h5 files;
Where EpsilonInfo stores the dielectric function data:

where the mechanical constant data is stored in ForceConstant:

Where PrimitiveAtomInfo, SupercellAtomInfo, and unitAtomInfo store the structural information for the primitive cell, supercell, and unit cell, respectively. Taking the unit cell as an example, the structure is as follows:
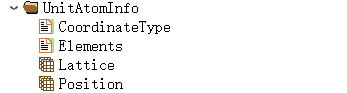
In which ThermalInfo stores phonon thermodynamic data:
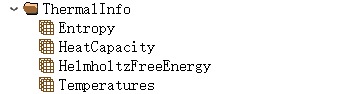
Here, Phonon stores the input parameters data for phonon calculations:
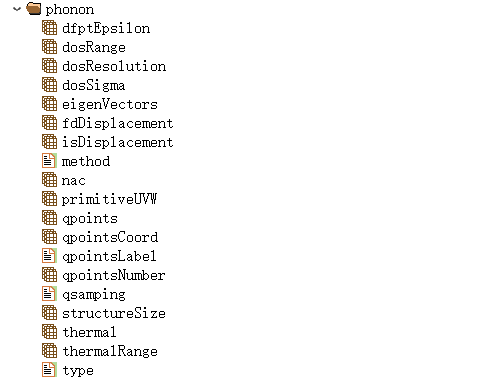
When
phonon.method = fd, the phonon.f5 file is shown as follows:
For phonon band structure and density of states calculations using the finite displacement method, phonon.h5 contains 9 structures:

Where BandInfo and DosInfo store band structure and density of states data, respectively, whose structures correspond to the structures in the files band.h5 and dos.h5, respectively.
where ForceConstant stores the force constant data, and PrimitiveAtomInfo, SupercellAtomInfo, and unitAtomInfo respectively
saving the structural information of the primitive cell, supercell, and unit cell, whose structures are consistent with the structures in the phonon.f5 file generated when phonon.method = dfpt;
Where ForceSet stores the mechanical matrix data calculated for each structure:

Here, the phonon calculation input parameter data is stored in Phonon:
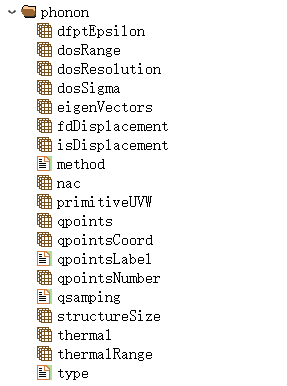
6.16 phonon001.h5
task = phonon, when phonon.method = fd, the file phonon.h5 will be output under the 001 subfolder. This type of h5 file can be renamed to phonon001.h5. Similarly, the phonon.h5 file will also be generated under the 002 folder.
In the case of spin consideration, phonon001.h5 contains 7 structures, as follows:

6.17 aimd.h5
aimd.h5 is the output file for task = aimd; it is not output when the task type is different.
When considering spin, aimd.h5 contains 9 structures:

The underlying structure information is consistent with relax.h5;
The new structure AimdInfo contains n structures, each storing the state information of the system under a certain ion step, such as temperature, pressure, energy, and kinetic energy:

6.18 epsilon.h5
epsilon.h5 is the output file for task = epsilon. This file is not produced for other task types
Considering spin, epsilon.h5 contains four structures:

In which the dielectric constant data is stored in EpsilonInfo:

6.19 wannier.h5
wannier.h5 is the output file under task = wannier; this file is not output when the task type is other than wannier.
Without considering spin, the interpolated bands calculated by wannier.h5 contain 9 structures:

In this, WannBandInfo stores the interpolated band data, which can be used to plot band diagrams:

Within this, WannInfo stores the Wannier function fitting data, including k-grid points and initial projection information, etc.:

In which, the spin1 under WannInfo stores the fitting data of Wannier functions, including the calculated Hamiltonian and other data:
